Joint Penn-UDel Seminar on DNA Computing
Efficient DNA Algorithms for Constrained Permutations for HPP, TSP, and ESD0
David H. Wood

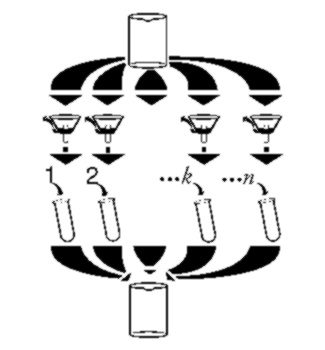
Extending partial permutations.
(Graphic by John Furno (c).)
Generating all possible permutations and then selecting
solutions is impractical. The number of permutations
n! is so great for a problem of interesting size that
an excessive amount of DNA is required.
The theme of stepwise building of only those permutations that
satisfy the problem constrains is illustrated.
Two methods are presented and applied to the HPP
and ESD0 problems.
The amount of work performed by the first method is Thursday, December 19, at 5pm (later than usual)
on the University of Pennsylvania campus,
in room 302 of the
Clinical Research Building
of the Medical School.
In many hard computational problems, the objective
is to find one or more permutations.
For example, an ordering of all the nodes of a graph,
allowing no duplications, solves HPP,
the Hamiltonian Path Problem.
The permutation property, an ordering of n objects
without duplications, is present in the solutions of many problems,
including
TSP,
the Traveling Salesman Problem,
BGM,
Matching in Bipartite Graphs,
and
ESD0,
Expansion of a Symbolic Determinant given its zero pattern.
work = O(n) sequential time steps
* O(n) lab steps done in parallel.
With the second method we have
work = O(n) sequential time steps
* O(n*n) lab steps done in parallel.
Both methods use only the quantities of DNA
required to encode the partial
permutations that may occur in the final answers.