
Joint Penn-UDel Colloquium on the Nature of Computing
Algorithmic Self-Assembly of DNA
Erik Winfree

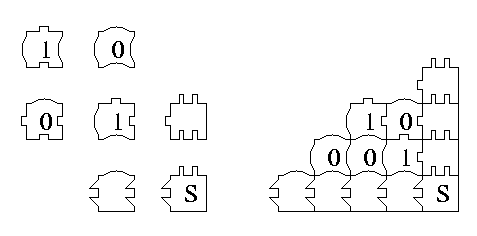
Fig. 1. Top: Tiles made of DNA. Bottom: Tiling Problem.
Biology makes things far smaller and more complex than anything produced by human engineering. The biotechnology revolution has for the first time given us the tools necessary to consider engineering on the molecular level. Research in DNA computation, launched by Len Adleman, has opened the door for experimental study of programmable biochemical reactions. This talk will focus on a single biochemical mechanism, the self-assembly of DNA structures, that is theoretically sufficient for Turing-universal computation. The theory combines Hao Wang's purely mathematical Tiling Problem with the branched DNA constructions of Nadrian Seeman. I will introduce branched DNA structures, illustrate the connection to the Tiling Problem, discuss biophysical issues that will determine error rates in any real system, and finally show experimental results confirming the self-assembly of a two-dimensional lattice of DNA Wang tiles. This system represents a first step toward the ability to program molecular reactions and molecular structures.