Joint Penn-UDel Colloquium on the Nature of Computing
Programming a Genetic Algorithm on a DNA Computer
David Harlan Wood

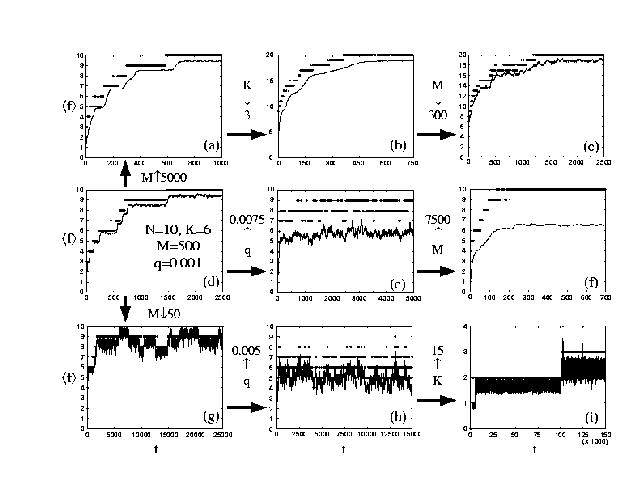
Fig. 1. Fitness evolution of a Genetic Algorithm for the "Royal Road" problem varies greatly with population size M, mutation rate q, and other parameters. (From Ref. 2, with permission.)
DNA computing techniques are desirable for Genetic Algorithm computations because they might process, in parallel, populations billions of times larger than is usual for conventional computers. However, selecting DNA strands for "breeding" is moderately challenging because one must physically separate DNA strands according to their "fitness." A DNA based Genetic Algorithm for a specific class of problems was first proposed last year (Ref. 1).
There are related methods in molecular biology referred to as "in-vitro evolution." Naturally, these methods use fitness criteria constrained to properties of biomolecular interest. In-vitro methods suit some very important but relatively small classes of problems seeking enzymes, ribozymes, binding sites, etc. Contrast this to genetic algorithms using unconstrained fitness criteria on bitstrings of length 100. Such computations can, in principal, evolve a population of fixed size in such a way as to create any one of 10^30 possible outcomes. Thus, Genetic Algorithms using DNA address larger, but less structured, classes of problems than does in-vitro evolution.
We are particularly interested in addressing the "Royal Road" problem using DNA. This problem got its name from the fact that it was intended to be especially suitable for Genetic Algorithms using crossover. Distressingly, computer trials exhibit a wide variety of unpleasant behaviors (see Fig. 1). A recent seminal paper (Ref. 2) attributes unanticipated behaviors for the Royal Road problem to limitations on population sizes. We hope to test the predictions of this paper using population sizes which are too large to be practical for conventional computers.
This research is joint work with Junghuei Chen, Eugene Antipov, Bertrand Lemieux, and Walter Cedeņo. Our group has obtained preliminary laboratory results using only conventional laboratory technology, including crossover based on a modified version of Stemmer's gene shuffling technique (Ref. 3).
1. Deaton, Murphy, Rose, Garzon, Franceschetti, and Stevens, "A DNA based implementation of an evolutionary search for good encodings for DNA computation," ICEC 1997, IEEE, 267-271.
2. Nimwegen, Crutchfield, and Mitchell, "Statistical dynamics of the Royal Road genetic algorithm," Theoretical Computer Science, to appear.
3. Stemmer, "Sexual PCR and Assembly PCR," in The Encyclopedia of Molecular Biology, Vol. 5, VCH, New York, 1996, 447-457.