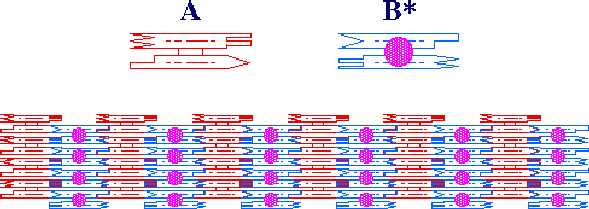
2-D arrays constructed from antiparallel double crossover molecules. These were produced by using two different double crossover molecules, which are shown at the top.
Joint Penn-UDel Colloquium on the Nature of Computing
DNA Nanotechnology
Nadrian C. Seeman

2-D arrays constructed from antiparallel double crossover molecules. These
were produced by using two different double crossover molecules,
which are shown at the top.
(General Interest: First 25 minutes)
DNA nanotechnology is the assembly of target molecules and materials from
unusual motifs of DNA, particularly branched molecules. These branched
motifs constitute a convenient system for the tractable assembly of
objects, knots and links on the nanometer scale. The addition of sticky
ends to branched DNA molecules converts them to convenient valence
clusters with addressable ends. In the past, we have assembled molecules
whose edges have the connectivities of a cube and a truncated octahedron
by using this approach. The assembly of periodic matter from these
components is a key goal of DNA nanotechnology. The minimal requirements
for components of designed crystals are
[1] programmable interactions,
[2]
predictable local intermolecular structures and
[3] rigidity.
The
sticky-ended association of DNA molecules fulfills the first two criteria,
because it is both specific and diverse, and because it results in the
formation of B-DNA. Stable branched DNA molecules permit the formation of
networks in principle, but simple branches are too flexible. Antiparallel
DNA double crossover (DX) molecules can provide the necessary rigidity, so
we can use these components to tile the plane.
(Break: Second 10 minutes)
(Researchers' Interests: Last 25 minutes)
It is possible to include DNA hairpins that act as topographic labels for this 2-D crystalline array, because they protrude out of the plane. By altering the sticky ends, it is possible to change the topographic features, and to detect these changes by means of Atomic Field Microscopy (AFM). We can modify the array by restricting hairpins or by adding them to the array.. Although simple 4-arm branched junctions are too flexible to be used as lattice components, parallelograms containing four single-crossover molecules are sufficiently rigid that they can be used to produce 2D arrays with tunable cavities.
The use of DNA suggests that self-reproduction might be feasible, perhaps by PCR. Unfortunately, it is not possible to reproduce a branched DNA molecule by means of DNA polymerases. Nevertheless, it is possible in principle to convert branched systems to single-stranded molecules that could be reproduced by polymerases and designed to fold into complex knots that could be trimmed to produce polyhedral species. As a first step in this direction, we have produced a variety of DNA knots. We have used an RNA knot to demonstrate the existence of RNA topoisomerase activity. Borromean rings are the most complex topological target we have constructed.
The rigidity of the DX motif can be exploited to produce a nanomechanical device predicated on the B-Z transition: This effect is shown by fluorescence energy resonance transfer, because two attached dyes have different separations in the two states. The system can be cycled between the two states.