Joint Penn-UDel Colloquium on the Nature of Computing
DNA Chip Technology and Genotyping
Bertrand R. Lemieux

|

|
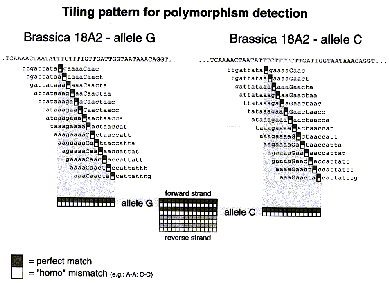
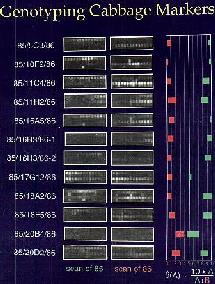
| Figure 1. Tiling strategy | Figure 2. Example data. |
Deoxyribonucleic acid (DNA) chip technology exploits high-density microarrays to quantitatively characterize the chemical composition of nucleic acid mixtures by hybridization of the components of these mixtures to the chips. Microarrays can be grouped into two categories based on the strategy that is used for their preparation. Microarrays belonging to the first class are made by the stepwise synthesis of oligonucleotides from nucleotide precursors, while the second class is prepared by the micro-deposition of pre-synthesized substances such as complementary DNAs (cDNAs). The manufacturing technologies used to make oligonucleotide and cDNA microarrays are as diverse as the research applications for which they are used.
We have used DNA chips as a platform for high-throughput genotyping. A set of single nucleotide polymorphisms (SNP) were identified by screening expressed sequence tag sites in Brassica napus and Brassica oleracea varieties. Two strategies can be used for polymorphism discovery (1) brute force sequencing or (2) heteroduplex analysis by denaturing high pressure liquid chromatography.
The 20 nucleotides that flank SNP markers were used to develop a tiling strategy. An example of the tiling strategy we used is given in Figure 1. The principle behind this strategy is that heteroduplexes with mismatches are less stable than perfectly matched heteroduplexes. Four strips of 14 oligonucleotides are used to genotype each strand. The gray boxes in Figure 1 represent perfect match primers for each allele while the white boxes are purposeful homo-mismatch oligonucleotides. The mismatch is introduced into the central nucleotide of the latter such as to maximize the destabilizing effect of the mismatch. Figure 2 is an example of the data we can extract with these genotyping DNA chips. The match and mismatch blocks are compared to estimate the probability that a given allele is being called. The graph to the right of the hybridization data is a crude representation of the output of the genotyping software.
Thursday, February 19, at 3 pm
in Room 207 of the
Johnson Pavilion
(see map)
near Spruce and 36th Streets on the
University of Pennsylvania campus.
Travel Directions are found at
http://www.upenn.edu/fm/map/dir.html